Evolution fills the functional gap
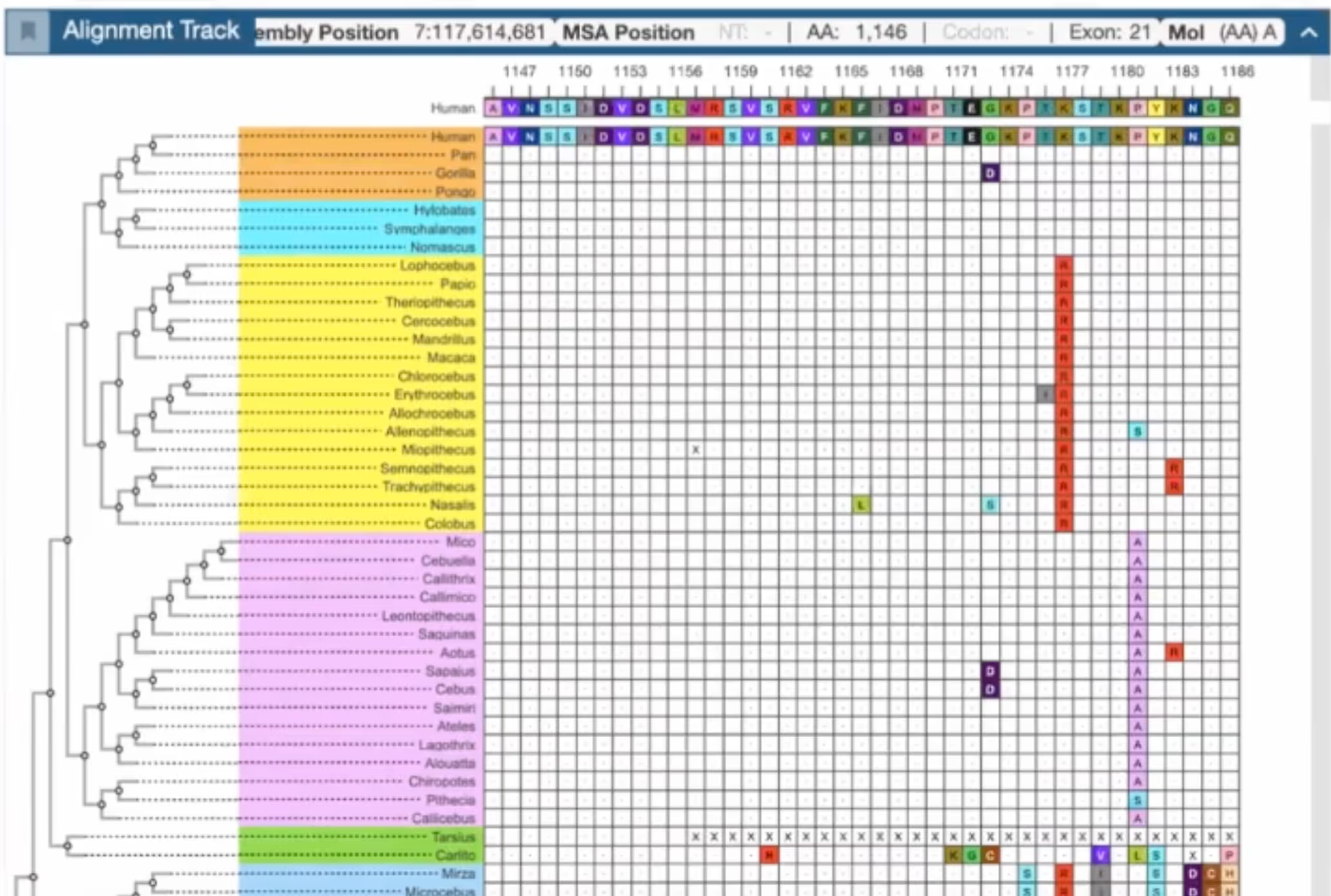
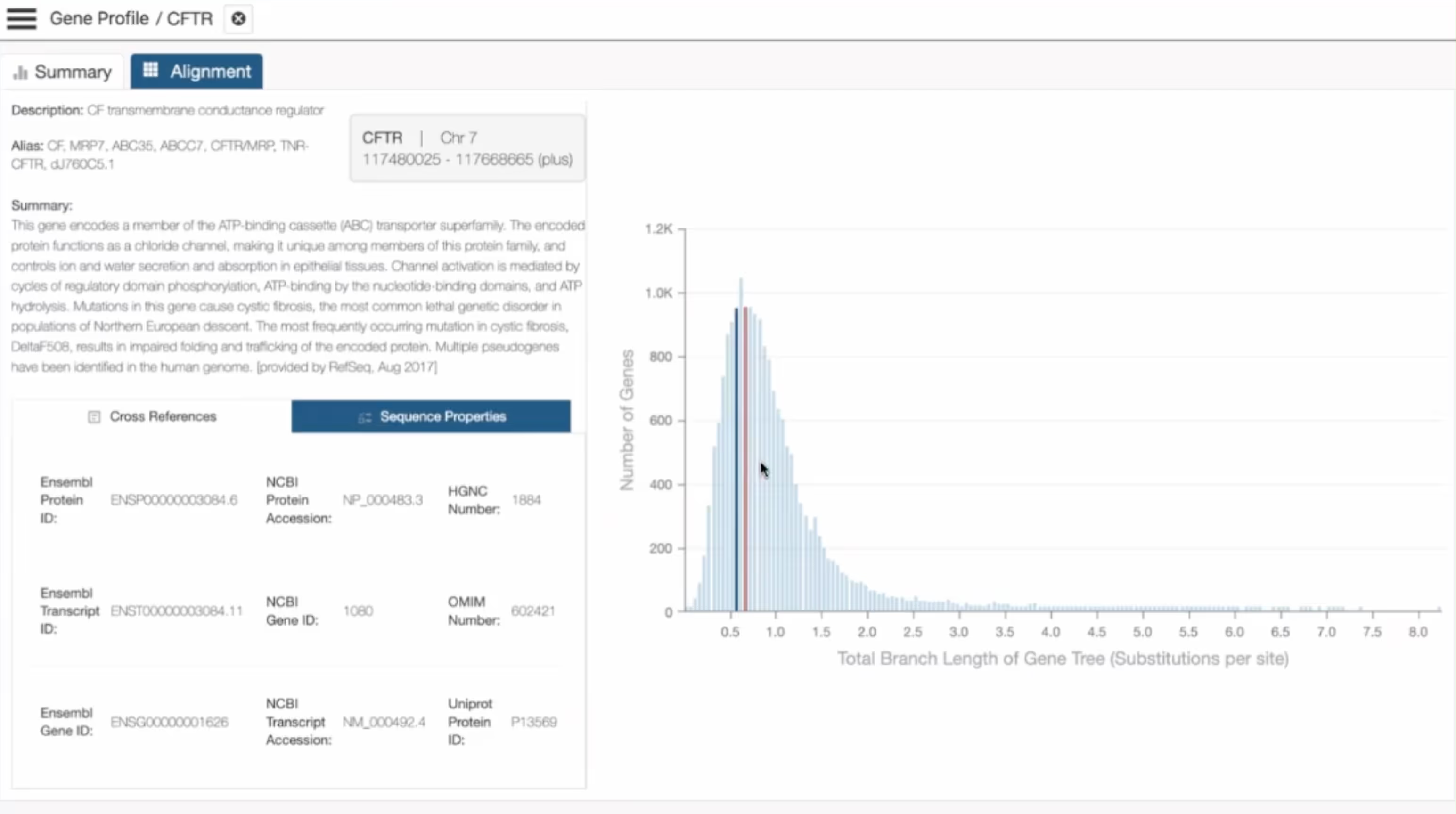
Deep-time recurrence shows which amino-acid substitutions biology tolerated across 55 primate lineages, revealing functional limits that never appear in human-only cohorts.
Deep-time evidence scales with your workload
Whether you are reviewing a single VUS or thousands of exome calls, CodeXome applies the same evolutionary constraints across your entire gene list in seconds.