Have you ever considered that genes don't all change at the same rate? This fundamental aspect of genetics is often overlooked, even among experts. Yet, understanding the varying rates at which genes evolve is critical for identifying disease-related genes and comprehending their impacts on human health.
The Basics of Gene Variation
Gene Changes Over Time
Genes evolve over time, accumulating changes at varying rates. Some genes remain highly conserved, while others evolve rapidly. This differential evolution is not just a biological curiosity; it plays a pivotal role in how we identify and interpret genes associated with diseases. By understanding these rates of change, we can better predict which genes might contribute to conditions like cancer, neurodegenerative diseases, or other genetic disorders.
Fast vs. Slow Regions
Within each gene, some regions change more rapidly than others. These fast and slow regions provide critical insights into the gene's function and its role in disease. For example, the ADCYAP1R1 gene, which plays a key role in the neuroendocrine stress response, exemplifies this concept. This gene encodes a membrane-bound receptor that initiates the stress response in the central nervous system (CNS). The gene's structure includes seven transmembrane loops, alternating between extracellular, transmembrane, and cytoplasmic regions.
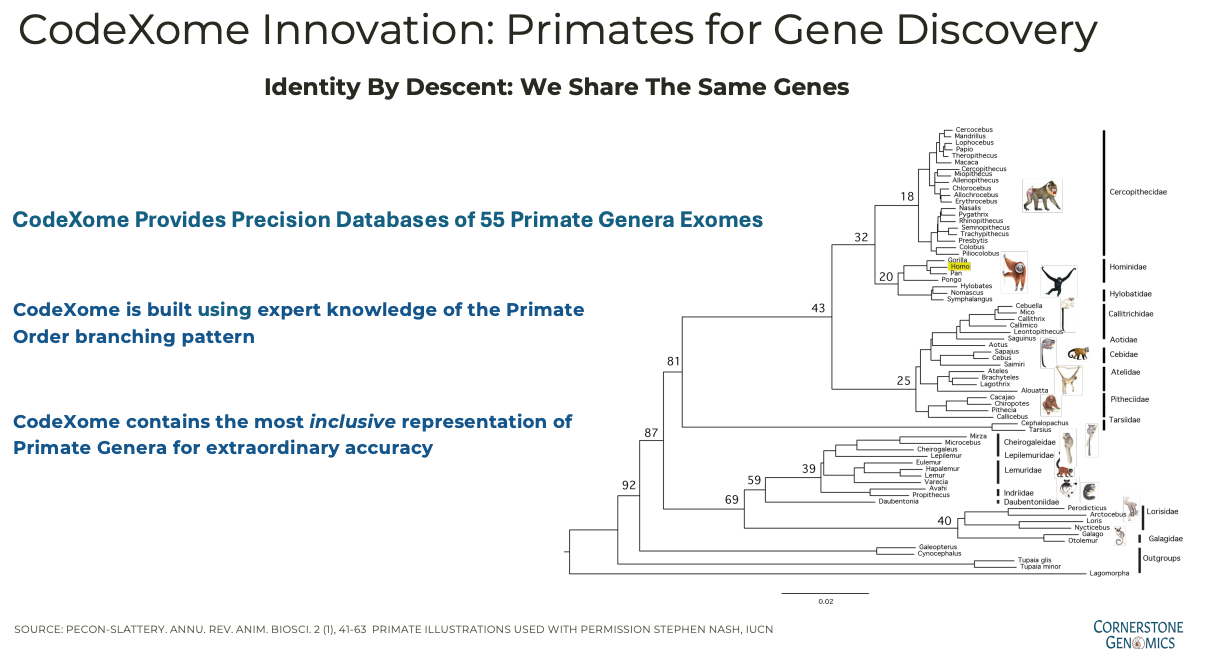
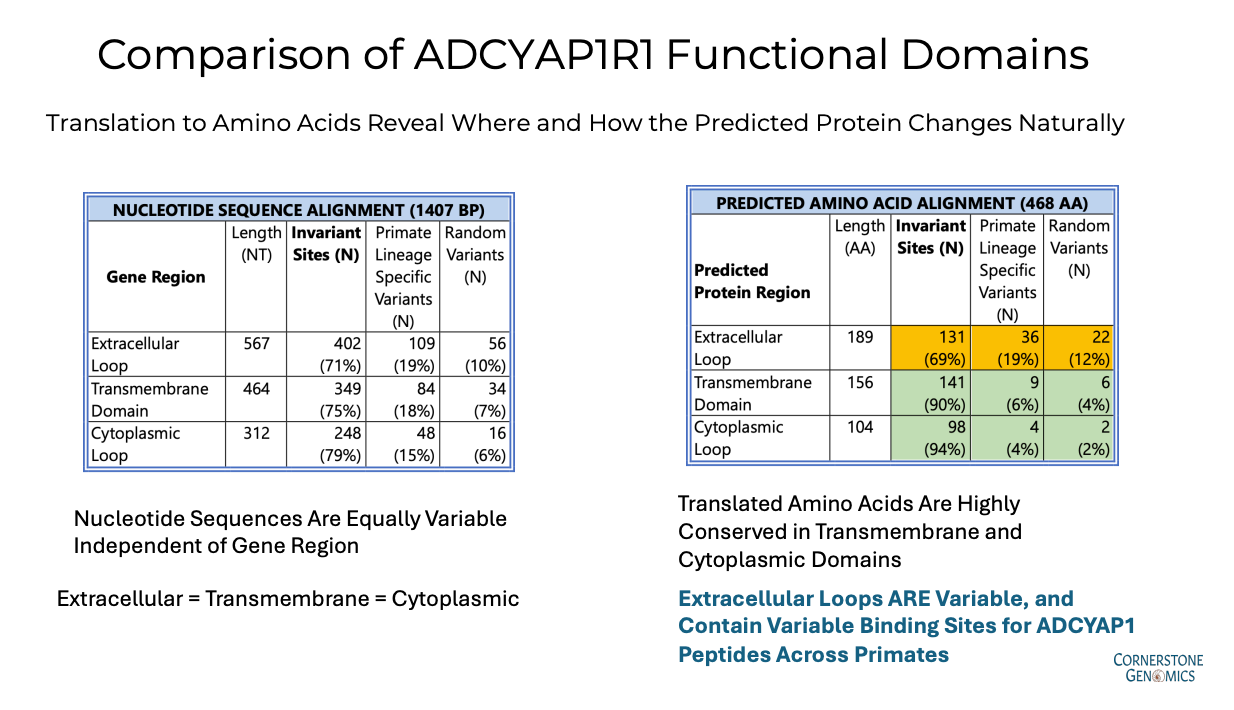
At Cornerstone Genomics, we sequenced ADCYAP1R1 across 55 primate genera, organizing the results according to distinct taxonomic families, such as Hominidae (Humans and Great Apes) and Hylobatidae (Gibbons). This extensive analysis, spanning 80 million years of evolution, reveals how each region of the gene and its encoded protein varies depending on its functional domain. For instance, nucleotide variation is slightly higher in extracellular regions (71% invariant) compared to transmembrane (75% invariant) and cytoplasmic regions (79% invariant).
Cornerstone Genomics and CodeXome Innovation: Primates for Gene Discovery
At Cornerstone Genomics, we’ve harnessed the power of evolutionary genomics to advance our understanding of genes linked with diseases. Our unique approach, embodied in the CodeXome platform, offers a precision database of exomes from 55 primate genera. This allows us to see how genes like ADCYAP1R1 evolve across different primate species and uncover the functional constraints that shape their evolution.
In the case of ADCYAP1R1, the true differences in function are revealed when the gene is translated into its predicted protein structure. The transmembrane and cytoplasmic regions of the receptor are highly conserved (90% and 94% invariant, respectively), indicating that observed nucleotide variation is mostly synonymous. By contrast, the extracellular region is less functionally constrained, allowing for more amino acid changes, particularly in the ligand-binding sites for the PACA peptides.
This differential evolution makes sense when considering the roles of these regions: while the extracellular regions interact with external ligands, the transmembrane and cytoplasmic regions are vital for maintaining the receptor's structural integrity and activating the cell's response to stress signals.
From Genes to Proteins
Genetic Signal Translation
Genetic information within a gene is translated into amino acids, forming proteins that are the functional molecules directly impacting a patient's condition. The initial genetic signal translates into a predicted protein structure, providing vital clues about the variant's impact.
Impact of Variants
Variants within a gene can alter the protein structure and function, with the location of the variant determining its impact. For instance, a variant in a highly conserved region like the transmembrane or cytoplasmic domains of ADCYAP1R1 is likely to have significant functional consequences, potentially leading to diseases like PTSD or other neurological disorders. On the other hand, variants in the more variable extracellular regions may have a subtler impact, possibly affecting ligand binding without disrupting the overall protein function.
Our Approach to Gene Analysis
Multi-Level Understanding
Our approach to gene analysis involves examining genes at multiple levels, combining evolutionary data with structural insights. This detailed, gene-specific analysis is far more effective than broad genome-wide approaches, as it uncovers the unique story of each gene. By focusing on the evolutionary narrative of individual genes, we provide a nuanced understanding that is crucial for interpreting genetic variants.
The Unique Story of Each Gene
Every gene has its own evolutionary story, which provides context for understanding its variants. For example, the KRAS gene, involved in many cancers, is one of the slowest-evolving genes, with almost no natural changes over 80 million years. This extreme conservation means that any substitution within KRAS can have a significant impact, often leading to cancer.
In contrast, sensory and immune genes evolve rapidly to adapt to environmental changes. These genes exhibit high rates of change and are involved in responses to external stimuli, highlighting the importance of context when interpreting genetic variants.
Practical Implications for Genetic Research
Identifying Disease-Related Genes
Understanding the rates of gene change is not just an academic exercise; it has practical implications for genetic research. By providing context for variant interpretation, we can more accurately identify genes involved in diseases and predict their impact on patient health.
Validating Functional Impacts
Validating the functional impacts of genetic variants requires a detailed analysis at both the gene and protein levels. This approach ensures accurate interpretation and better-informed medical interventions. For example, understanding the tight evolutionary constraints on the transmembrane and cytoplasmic regions of ADCYAP1R1 can help researchers design more effective therapies for PTSD and other neurological disorders.
Conclusion
Understanding gene variation rates and their implications is crucial for advancing genetic research and improving patient outcomes. We believe that researchers and clinicians should consider this detailed approach in their work to uncover new insights and develop more effective treatments. The rich tapestry of genetic information holds the potential to unlock groundbreaking medical discoveries, emphasizing the importance of detailed and contextual analysis.
By delving into the intricacies of gene evolution, we can better understand the genetic underpinnings of diseases and pave the way for more precise and personalized medical interventions.




